Chlorine »
PDB 7vkh-7vzz »
7vyq »
Chlorine in PDB 7vyq: Short Chain Dehydrogenase (Scr) Cryoem Structure with Nadp and Ethyl 4-Chloroacetoacetate
Chlorine Binding Sites:
The binding sites of Chlorine atom in the Short Chain Dehydrogenase (Scr) Cryoem Structure with Nadp and Ethyl 4-Chloroacetoacetate
(pdb code 7vyq). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the Short Chain Dehydrogenase (Scr) Cryoem Structure with Nadp and Ethyl 4-Chloroacetoacetate, PDB code: 7vyq:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the Short Chain Dehydrogenase (Scr) Cryoem Structure with Nadp and Ethyl 4-Chloroacetoacetate, PDB code: 7vyq:
Jump to Chlorine binding site number: 1; 2; 3; 4;
Chlorine binding site 1 out of 4 in 7vyq
Go back to
Chlorine binding site 1 out
of 4 in the Short Chain Dehydrogenase (Scr) Cryoem Structure with Nadp and Ethyl 4-Chloroacetoacetate

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of Short Chain Dehydrogenase (Scr) Cryoem Structure with Nadp and Ethyl 4-Chloroacetoacetate within 5.0Å range:
|
Chlorine binding site 2 out of 4 in 7vyq
Go back to
Chlorine binding site 2 out
of 4 in the Short Chain Dehydrogenase (Scr) Cryoem Structure with Nadp and Ethyl 4-Chloroacetoacetate
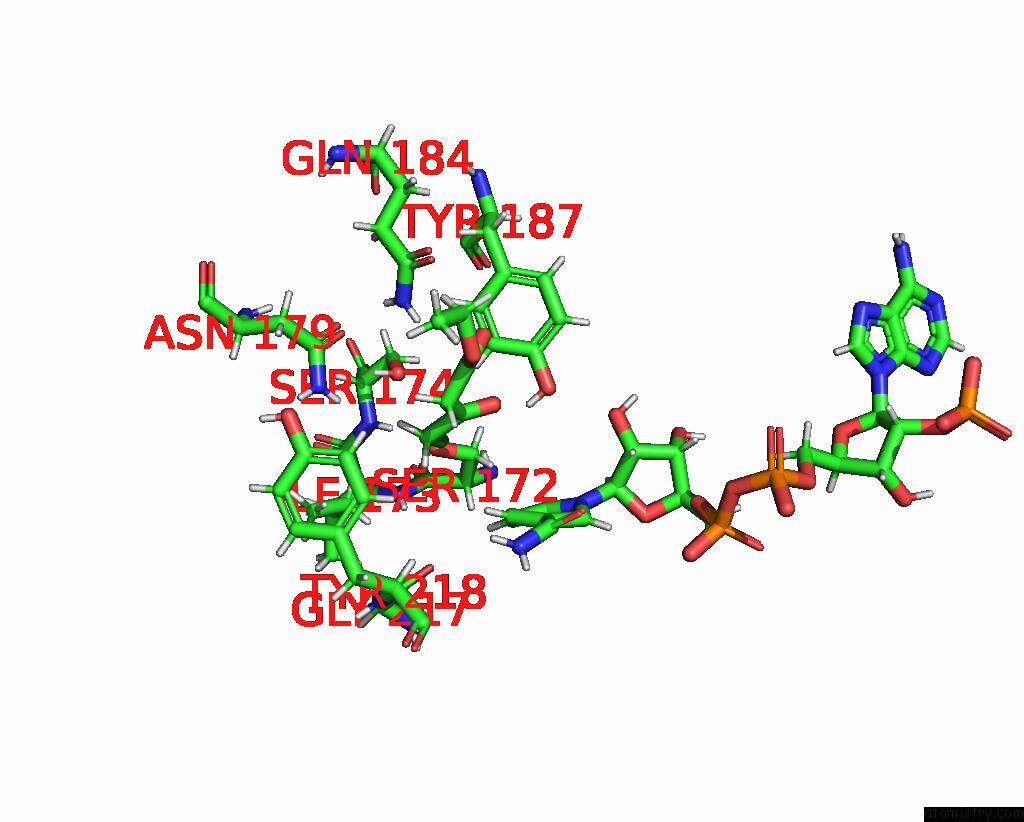
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of Short Chain Dehydrogenase (Scr) Cryoem Structure with Nadp and Ethyl 4-Chloroacetoacetate within 5.0Å range:
|
Chlorine binding site 3 out of 4 in 7vyq
Go back to
Chlorine binding site 3 out
of 4 in the Short Chain Dehydrogenase (Scr) Cryoem Structure with Nadp and Ethyl 4-Chloroacetoacetate
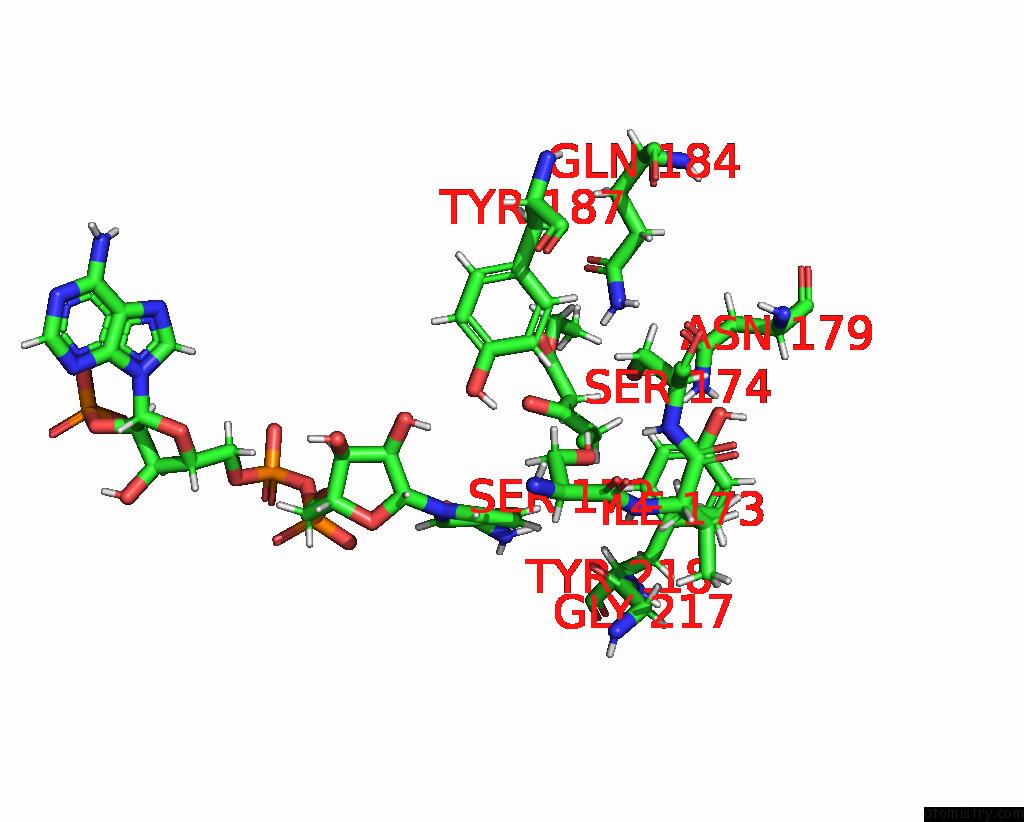
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of Short Chain Dehydrogenase (Scr) Cryoem Structure with Nadp and Ethyl 4-Chloroacetoacetate within 5.0Å range:
|
Chlorine binding site 4 out of 4 in 7vyq
Go back to
Chlorine binding site 4 out
of 4 in the Short Chain Dehydrogenase (Scr) Cryoem Structure with Nadp and Ethyl 4-Chloroacetoacetate
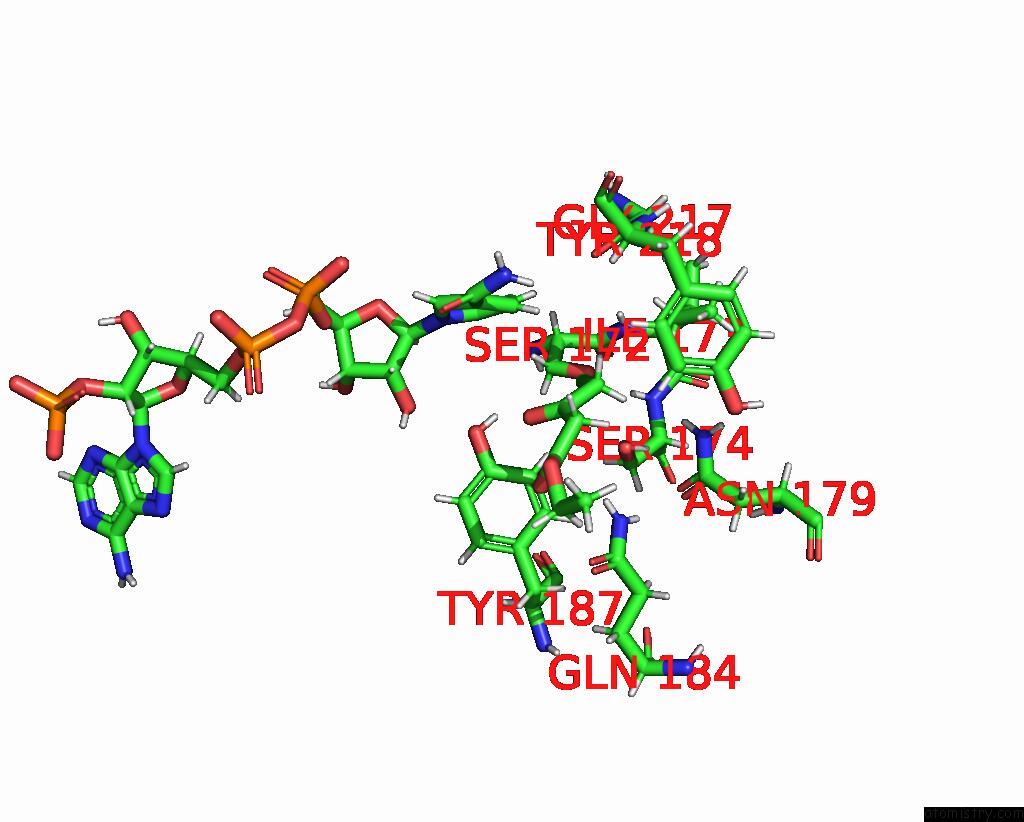
Mono view
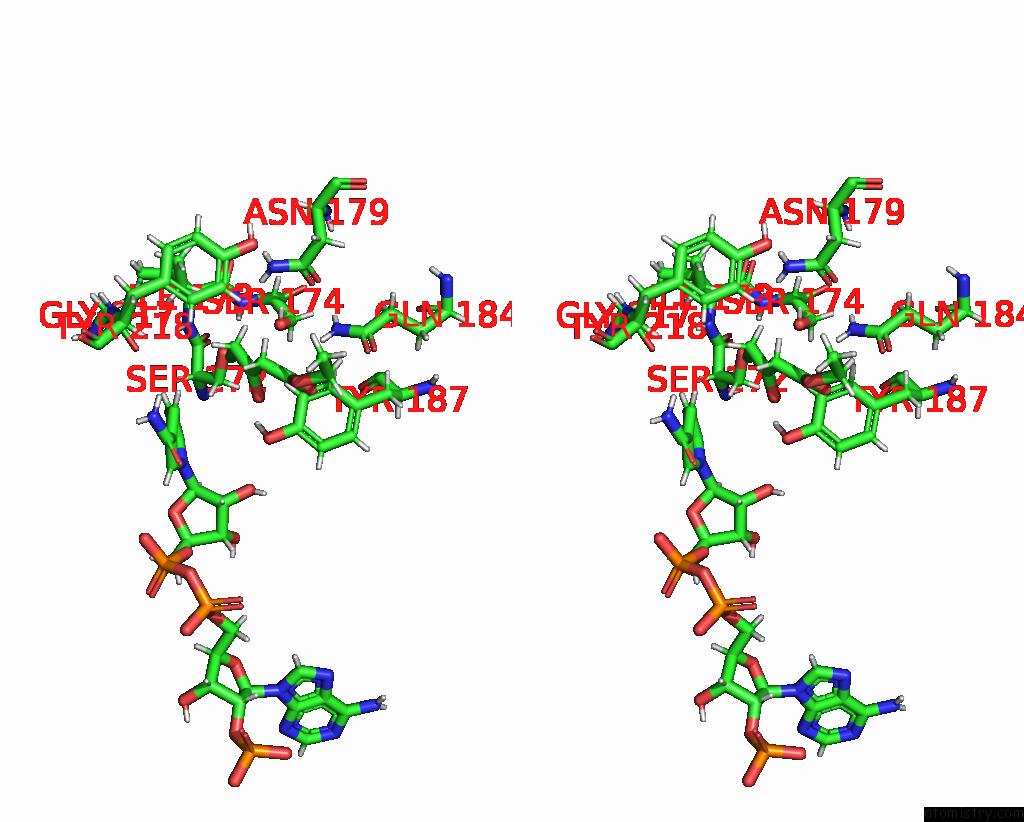
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of Short Chain Dehydrogenase (Scr) Cryoem Structure with Nadp and Ethyl 4-Chloroacetoacetate within 5.0Å range:
|
Reference:
Y.Li,
R.Zhang,
C.Wang,
F.Forouhar,
O.B.Clarke,
S.Vorobiev,
S.Singh,
G.T.Montelione,
T.Szyperski,
Y.Xu,
J.F.Hunt.
Oligomeric Interactions Maintain Active-Site Structure in A Noncooperative Enzyme Family. Embo J. V. 41 08368 2022.
ISSN: ESSN 1460-2075
PubMed: 35801308
DOI: 10.15252/EMBJ.2021108368
Page generated: Sun Jul 13 08:12:29 2025
ISSN: ESSN 1460-2075
PubMed: 35801308
DOI: 10.15252/EMBJ.2021108368
Last articles
Cl in 9DSBCl in 9DW9
Cl in 9DQK
Cl in 9DTV
Cl in 9DW6
Cl in 9DUJ
Cl in 9DQL
Cl in 9DSN
Cl in 9DRX
Cl in 9DR6