Chlorine »
PDB 1eyz-1g0g »
1fu2 »
Chlorine in PDB 1fu2: First Protein Structure Determined From X-Ray Powder Diffraction Data
Other elements in 1fu2:
The structure of First Protein Structure Determined From X-Ray Powder Diffraction Data also contains other interesting chemical elements:
| Zinc | (Zn) | 4 atoms |
| Sodium | (Na) | 2 atoms |
Chlorine Binding Sites:
The binding sites of Chlorine atom in the First Protein Structure Determined From X-Ray Powder Diffraction Data
(pdb code 1fu2). This binding sites where shown within
5.0 Angstroms radius around Chlorine atom.
In total 4 binding sites of Chlorine where determined in the First Protein Structure Determined From X-Ray Powder Diffraction Data, PDB code: 1fu2:
Jump to Chlorine binding site number: 1; 2; 3; 4;
In total 4 binding sites of Chlorine where determined in the First Protein Structure Determined From X-Ray Powder Diffraction Data, PDB code: 1fu2:
Jump to Chlorine binding site number: 1; 2; 3; 4;
Chlorine binding site 1 out of 4 in 1fu2
Go back to
Chlorine binding site 1 out
of 4 in the First Protein Structure Determined From X-Ray Powder Diffraction Data
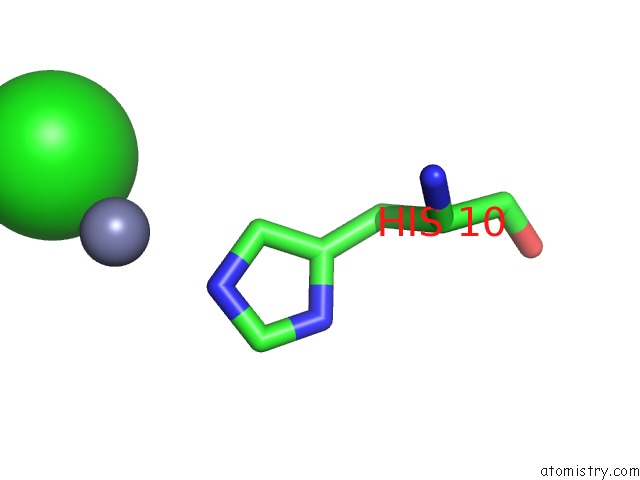
Mono view
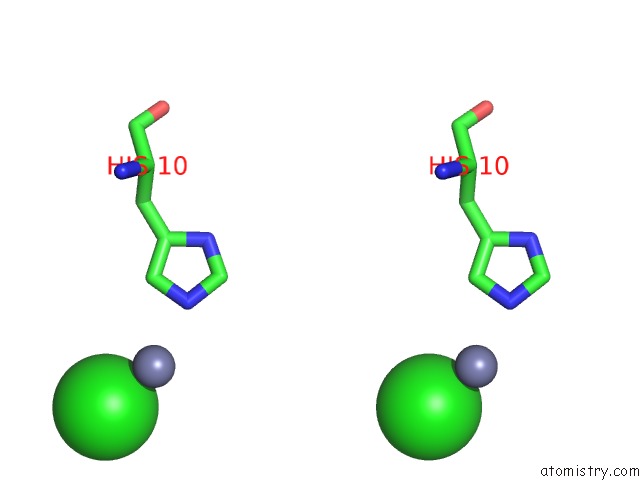
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 1 of First Protein Structure Determined From X-Ray Powder Diffraction Data within 5.0Å range:
|
Chlorine binding site 2 out of 4 in 1fu2
Go back to
Chlorine binding site 2 out
of 4 in the First Protein Structure Determined From X-Ray Powder Diffraction Data
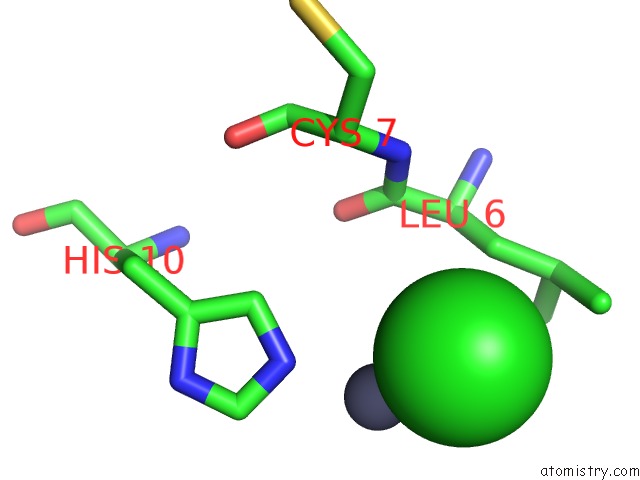
Mono view
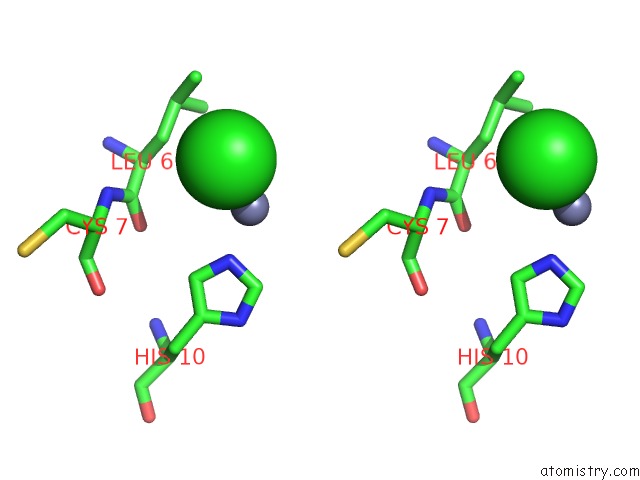
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 2 of First Protein Structure Determined From X-Ray Powder Diffraction Data within 5.0Å range:
|
Chlorine binding site 3 out of 4 in 1fu2
Go back to
Chlorine binding site 3 out
of 4 in the First Protein Structure Determined From X-Ray Powder Diffraction Data
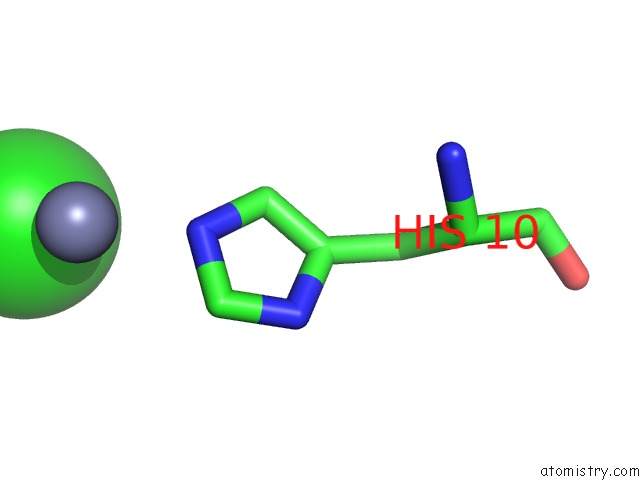
Mono view
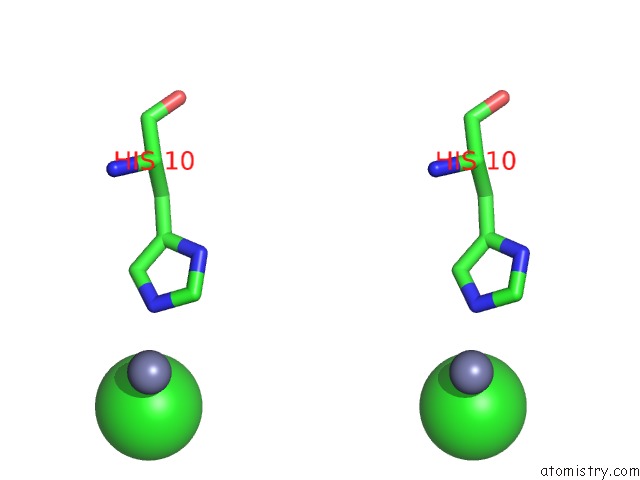
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 3 of First Protein Structure Determined From X-Ray Powder Diffraction Data within 5.0Å range:
|
Chlorine binding site 4 out of 4 in 1fu2
Go back to
Chlorine binding site 4 out
of 4 in the First Protein Structure Determined From X-Ray Powder Diffraction Data
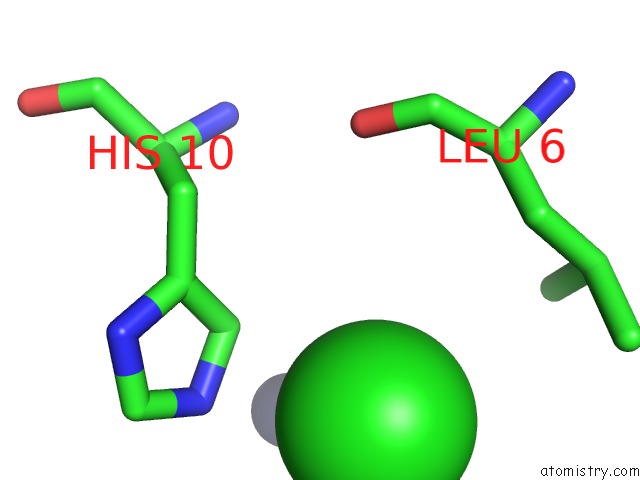
Mono view
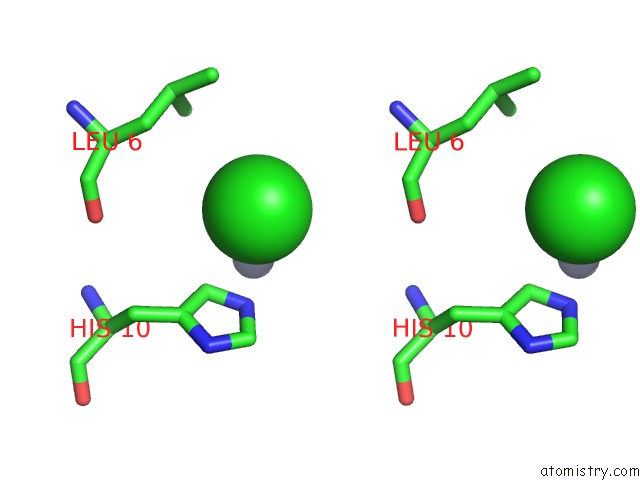
Stereo pair view

Mono view

Stereo pair view
A full contact list of Chlorine with other atoms in the Cl binding
site number 4 of First Protein Structure Determined From X-Ray Powder Diffraction Data within 5.0Å range:
|
Reference:
R.B.Von Dreele,
P.W.Stephens,
G.D.Smith,
R.H.Blessing.
The First Protein Crystal Structure Determined From High-Resolution X-Ray Powder Diffraction Data: A Variant of T3R3 Human Insulin-Zinc Complex Produced By Grinding. Acta Crystallogr.,Sect.D V. 56 1549 2000.
ISSN: ISSN 0907-4449
PubMed: 11092920
DOI: 10.1107/S0907444900013901
Page generated: Thu Jul 10 16:54:43 2025
ISSN: ISSN 0907-4449
PubMed: 11092920
DOI: 10.1107/S0907444900013901
Last articles
Cl in 5UGSCl in 5UJS
Cl in 5UHI
Cl in 5UJK
Cl in 5UJ2
Cl in 5UIN
Cl in 5UI2
Cl in 5UGT
Cl in 5UHR
Cl in 5UGH